Abstract
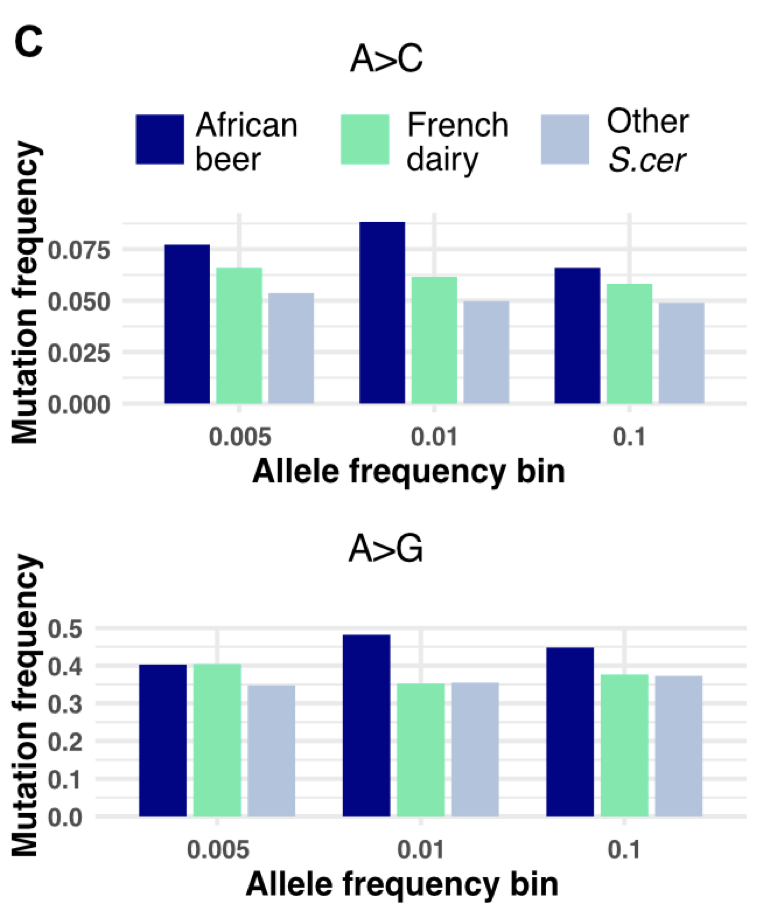
Most mutations are neutral or deleterious, and mutator alleles that increase the mutation rate of an organism are considered rare and short-lived. Here, we report a genomic signature consistent with the transmission and long-term maintenance of natural mutator allele(s) in Saccharomyces cerevisiae isolates. Specifically, we identified genomic signatures of standing mutator allele(s) that disproportionately increase A>C and A>G mutations in natural polymorphisms of the African beer population of S. cerevisiae. Remarkably, the mutation spectrum deviation in this population is greater than that observed between some Saccharomyces species. Furthermore, computational analysis suggests the introgression of mild mutator allele(s) from the African beer population into a subset of the French dairy population, motivating experimental characterization of de novo mutations in these strains. We observed a consistent but weak enrichment of A>C and A>G mutations among de novo mutations in the African beer population, likely reflecting their low overall frequency. Other mutation types showed greater variability, with one outlier strain (AFL from African beer) exhibiting an excess of C>A mutations. Comparisons across strains that have been empirically assayed for mutation spectra revealed that de novo mutations display the greatest variability, followed by rare polymorphisms. This pattern suggests that additional mutator alleles may segregate in natural populations but are often purged due to their fitness costs. We infer that the mild enrichment of A>C and A>G mutations in African beer strains reflects mutator alleles with weak effects that can persist through evolution.